Molecular Immunology: Antigen Processing and Presentation in Human Dendritic Cells
One of the vital interactions for the adaptive immune response against pathogens or malignant transformed cells is between antigen loaded major histocompatibility complex class I (MHC I) molecules and CD8-positive T-cells. The transporter associated with antigen processing (TAP) together with MHC I and other chaperones, known as the peptide loading complex (PLC), build the core in the classical MHC I antigen presentation pathway. Endogenous peptides, derived from proteasomal degradation, are translocated by TAP into the endoplasmatic reticulum (ER) lumen and consequently loaded onto MHC I molecules by the PLC to elicit successful cytotoxic responses by T-cells. Professional antigen-presenting cells, such as dendritic cells (DCs), take antigen presentation one step further, by cross-presenting exogenous peptides on MHC I molecules. However, the key characteristics of this mechanism remain to be discovered.
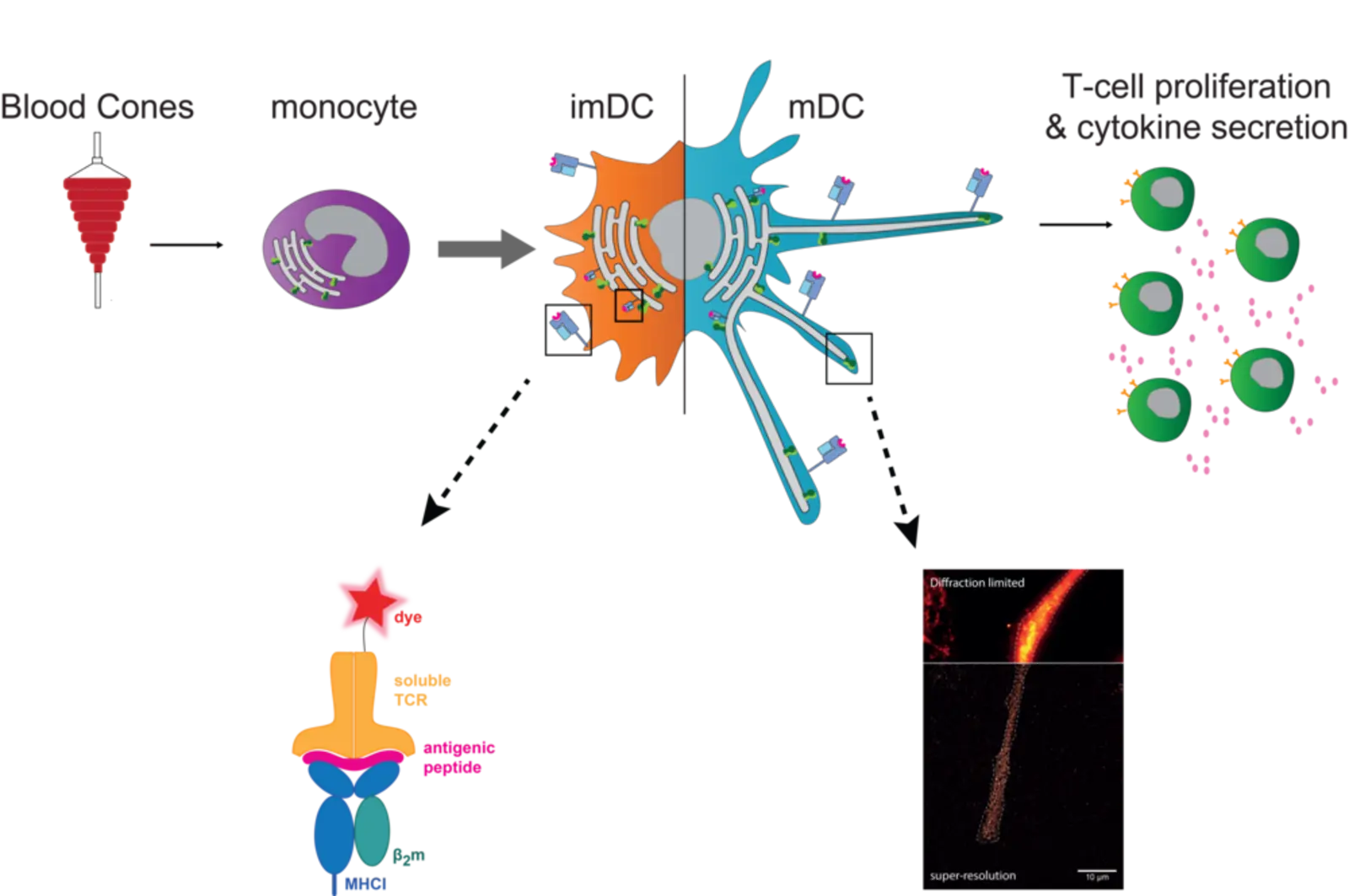
In our molecular immunology team, primary monocyte-derived DCs are used to follow the fate of PLC. Traced by TAP, along the maturation process from monocytes to DCs, a combination of classical biochemical and cell biological approaches is used - such as CRISPR/Cas9 knockouts together with state-of-the-art super-resolution microscopy. As immunogenic peptides are a key component of the interaction between dendritic- and T-cells, we also focus on the intra- and extracellular visualization of specific immune antigens by soluble T-cell receptors and click-chemistry.
Tackling the MHC I Peptide-Loading Complex in Human Dendritic Cells
In the classical MHC I antigen presentation pathway, the PLC allows the transport of cytosolic peptides, derived from proteasomal degraded endogenous proteins, into the ER lumen and consequently MHC I-peptide loading. In cross-presentation, exogenously acquired antigens are displayed by MHC I as well. For this, DCs are considered to play a key role in vivo. Overall, the exact mechanisms of antigen presentation are not clear yet and therefore still subject to investigation. In monocytes, the PLC traced by TAP is mainly located in early endosomes. These cells are highly plastic and an further differentiate to DCs. After differentiation from monocytes to immature DCs (imDCs), TAP re-locates to a perinuclear distribution along with the ER marker calnexin. Upon further maturation to mature DCs (mDCs), these cells elongate and the PLC is re-distributed towards the tips of these dendrites, where it accumulates to a high degree. Whether the re-localization of the PLC to the tips of human mDCs is associated with a change in the site of antigen processing and MHC I peptide loading, remains to be answered. For this, visualization of the intra- and extracellular antigen fate is aimed using labeled antigens and soluble T-cell receptors. Hence, to tackle these questions, we work with cell lines and human primary cells and apply diverse methods ranging from cell culture, knockouts by CRISPR/Cas9, T-cell stimulation assays to flow cytometry, biochemical approaches and super-resolution microscopy.
In summary, to further comprehend the degree of PLC dependency in MHC I antigen presentation and its impact on T-cell activation, we aim to i) investigate the modulation of the PLC and its influence on antigen presentation capacity in DCs and ii) visualize MHC I antigen presentation pathways.
Publications:
Döring M, Blees H, Koller N, Tischer-Zimmermann S, Musken M, Henrich F, Becker J, Grabski E, Wang J, Janssen H, Zuschratter W, Neefjes J, Klawonn F, Eiz-Vesper B, Tampé R, Kalinke U
Modulation of TAP-dependent antigen compartmentalization during human monocyte-to-DC differentiation.
2019; Blood Adv,3, 839-850, doi: 10.1182
Braner M, Koller N, Knauer J, Herbring V, Hank S, Wieneke R, Tampé R
Optical control of the antigen translocation by synthetic photo-conditional viral inhibitors.
2018; Chem Sci, 10, 2001-5; doi: 10.1039/c8sc04863k
Blees A, Januliene D, Hofmann T, Koller N, Schmidt C, Trowitzsch S, Moeller A, Tampé R
Structure of the human MHC-I peptide-loading complex.
2017, Nature, 551, 525-8; doi: 10.1038/nature 24627
- Cover Story and News&View in Nature 551,442-3
- Dispatch in Curr Biol 28,R83-R86
- Highlight in Faculty 1000 Prime
Fischbach H, Döring M, Nikles E, Lehnert C, Baldauf U, Kalinke U, Tampé R
Ultrasensitive quantification of TAP-dependent antigen compartmentalization in scarce primary immune cell subsets.
2015; Nat Commun,6, 6199; doi: 10.1038/ncomms7199
Project members
Olivia Jacobs
PhD Student
phone +49-(0)69-798-29505
o.jacobs@biochem.uni-frankfurt.de
Miriam Albensoeder
PhD Student
phone +49-(0)69-798-29505
m.albensoeder@biochem.uni-frankfurt.de